| Journal of Hematology, ISSN 1927-1212 print, 1927-1220 online, Open Access |
| Article copyright, the authors; Journal compilation copyright, J Hematol and Elmer Press Inc |
| Journal website http://www.thejh.org |
Original Article
Volume 8, Number 3, September 2019, pages 111-120
Quantitative Assay of Mutated Nucleophosmin in Acute Myeloid Leukemia
Safaa A.A. Khaleda, b, e, John Burthemc, El-Badry E. Abo Elnoora, Lobna F. ElTonia, Hanan M. Ahmeda, Sohier M. Ahmedd
aDepartement of Internal Medicine, Clinical Hematology Unit, Assiut University Hospital, Assiut, Egypt
bBone Marrow Transplantation Unit, South Egypt Cancer Institute, Assiut University, Assiut, Egypt
cCenter of Hematological Malignancies, Manchester University, Manchester, UK
dDepartment of Clinical Pathology and Laboratory Hematology, Assiut University Hospital, Assiut, Egypt
eCorresponding Author: Safaa A.A. Khaled, Department of Internal Medicine, Clinical Hematology Unit, Assiut University Hospital, Assiut 71526, Egypt
Manuscript submitted June 19, 2019, accepted August 9, 2019
Short title: Assay of Nucleophosmin in AML
doi: https://doi.org/10.14740/jh390
| Abstract | ▴Top |
Background: In our previous work, we provided strong evidence that nucleophosmin (NPM) gene mutation has an important role in leukemogenesis of primary acute myeloid leukemia (AML). Furthermore, we speculated a new targeted therapy in patients with primary AML and bearing mutated NPM (mNPM). Based on these results together with findings of other researchers, it was essential to develop a method for accurate detection of mNPM.
Methods: Our method based on utilizing the most recent flow cytometeric techniques and instruments in measuring mNPM. Attributed to their availability and technical feasibility, we used human leukemia cell lines to validate our method.
Results: The main findings were differential expression of wild-type NPM (wtNPM) within the same sample. Furthermore flow cytometry (FCM) was a simple straightforward tool for quantitative assay of mNPM.
Conclusions: In this work we developed an innovative technique that could enable quantitative assay of mNPM, and ease its use as a biomarker in cytogenetic and molecular prognostication of primary AML. In addition the study suggested that FCM could differentiate mNPM expression within cells of the same patient thus could be used for monitoring of minimal residual disease.
Keywords: Nucleophosmin; Assay; Quantitative
| Introduction | ▴Top |
The recent approach in treatment of patients with de novo acute myeloid leukemia (AML) depends on specific risk adapted treatment protocols. Cytogenetic and molecular criteria of AML were found to be the most important factors that determined the risk group for each individual patient. Nucleophosmin (NPM) gene mutation was speculated to be one of these factors. This was mainly valid in AML patients lacking other cytogenetic abnormalities [1-5]. Accordingly, several attempts were conducted to develop a suitable method for detection of mutated NPM (mNPM) in primary AML.
The first reported detection of mNPM in AML was based mainly on immunohistochemical (IHC) analysis to spot the cytoplasmically translocated NPM, followed by direct sequence analysis of leukemic DNA. For this purpose, Falini et al obtained a pretreatment bone marrow biopsy from patients enrolled in their study [6, 7]. This approach of NPM mutational analysis gave accurate results; however bone marrow biopsy is an invasive technique which requires an expert hand, besides it is not an integral part of the routine diagnostic/prognostic workup of AML. Moreover, direct sequence analysis is a time consuming, expensive procedure, which is attributed to its limit of detection of nearly 20% of a minor allele population, which necessitates cloning and sequencing of several clones. A recent study in 2016 reaffirmed the validity of IHC analysis in detection of mNPM and recommended this method in dry tap AML or myeloid sarcoma [8].
The above mentioned method was followed by several researches in succession to develop a reliable assay of mNPM in AML patients and ease its use as a molecular biomarker. In 2005, a group of Italian scientists described a method for screening NPM mutations by real-time polymerase chain reaction (RT-PCR) followed by denaturing high performance liquid chromatography. However this method is complex and depends on sophisticated equipments that are not available in many centers [9]. Calvo et al designed RT-PCR strategy to amplify NPM exon 12 followed by electrophoresis and fragment visualization on polyacrylamide gels in order to discriminate a 4 - 5 base pair size difference resulting from mutations in this gene [10]. Another PCR-based assay for NPM mutants has been devised, using differential melting of an oligo probe labeled with a fluorescent dye. The nucleobase quenching phenomenon was used to detect probe hybridization, and an oligonucleotide containing locked nucleic acid nucleotides was used as a PCR clamp to suppress amplification of the normal sequence and enhance the analytical sensitivity of the assay. The analytical sensitivity of this assay is variable depending on the concentration of the PCR clamp and other parameters [11]. Some researchers tried detection of type A mutation using fluorescence resonance energy transfer (FRET) probes [12]. However, none of these methods is a standard method for quantitative assay of mNPM; this was mainly due to the complex lengthy nature of these methods besides they are not available in all laboratories.
Other researchers tried to use flow cytometry (FCM) to detect mNPM in AML. Grimwade et al found that imaging FCM could identify mNPM in bone marrow samples of patients with AML. Also Du Pisani and Shires developed a flow cytometric technique that enables the identification of mNPM in AML patients. Nevertheless, false positive cases in their techniques, small sample size, and defective quantification indicate the need for further research [13, 14].
| Materials and Methods | ▴Top |
Cells, cell culture and antibodies
Both OCI-AML3 and HL60 purchased from DSMZ-German Collection of Cell Cultures (ACC 582,3) were grown in RPMI 1640 media, with GlutaMAX™ supplemented with 10% FCS and 1% Pen/strep (GIBCO, 61870- and 15070044, 10108-165, -063), at optimum gas and temperature conditions (37 °C, 5% CO2). Human leukemia cell lines HL60 and OCI-AML3 were used as a model for NPMc- and NPMc+ AMLs, respectively.
Primary antibodies mouse anti-NPM (Zymed, FC-61991) which reacts with the C-terminus of NPM (detects wtNPM only), and a polyclone to NPM (abcam, 15440) which reacts to amino acid residues 23 - 38 and 226 - 240 of human NPM (detects both wtNPM and mNPM) were used.
Secondary antibodies, goat anti-rabbit (abcam, ab5999) conjugated with fluorescein isothiocyanate (FITC), and goat anti-mouse (abcam, ab6003) conjugated with Texas Red (TR).
Flow cytometeric analysis
We carried out a flow cytometeric analysis of both OCI-AML3, and HL60 cells (cell line models for NPMc+ and NPMc- AMLs, respectively) labeled with anti-human NPM antibody that could recognize wtNPM only and another that recognizes both wtNPM and mNPM (ab15440 or anti-pan NPM). Fluorescence-activated cell sorting (FACS) analysis was performed with the BD FACS Canto II machine, equipped with BD FACS Diva v4.1.
We were guided by other researchers in interpreting flow cytometric data where FACS data were presented with logarithmic axes that extend over five decades [15]. Data were displayed as a single parameter frequency histogram that allows direct visual comparison, and a bivariate dot plot that helps detection of relative levels of other parameters displayed at the same time. NPM protein levels were represented with the mean fluorescence values of the correspondent anti-NPM antibody.
Specificity of the method was mainly dependent on the specificity of the anti-NPM antibody, while the accuracy was based on the flow cytometric potential in providing precise measurement of the cellular NPM-protein.
Immunocytochemistry (immunostaining)
Firstly cellular cytospins were prepared from living cells. Then, we followed the manufacturer recommended protocols for immunostaining of cytospins for wtNPM and pan -NPM. Next, the slides were viewed, with the Nikon Eclipse 80I fluorescence microscope (ICCT Technologies, Canada), equipped with three fluorescence filter cubes. The obtained images were grabbed with the Hamamatsu ORCA HR (C4742-95-12HR) high resolution digital camera, with full remote control from PC. Another fast high sensitivity black and white camera was used for low intensity fluorescence slides. Immunofluorescence images were captured, analyzed and saved with the NIS-Elements imaging software. Also, creation of overlay images (e.g. 4',6-diamidino-2-phenylindole (DAPI)/NPM overlay) was carried out with the same program. The saved images were edited with the Adobe Photoshop or Picasa programs.
Ethical considerations
The study protocol and objectives were consistent with the Declaration of Helsinki for medical research; furthermore the protocol was approved by the Institutional Review Board of Faculty of Medicine, Assiut University.
| Results | ▴Top |
FCM is a simple straightforward tool for quantitative assay of mNPM
In order to assess the practicability of FCM in measuring mNPM, a minimum of 1 × 106 of HL60 and OCI-AML3 cells were fixed, then permeabilized, and next incubated with the primary NPM antibody. The antigen antibody reactions were detected with the use of species specific phycoerythrin (PE)-conjugated secondary antibodies, while the negative controls were stained with the secondary antibody only. Details of the method were discussed previously.
FACS analysis of OCI-AML3 labeled with anti-pan NPM showed single peak MF extending beyond the fourth decade, with 99.9% of total OCI-AML3 population expressing total NPM compared with 99.6% only in HL60. Although OCI-AML3 negative controls showed much more non-specific bindings than the HL-60 ones, total NPM MF of OCI-AML3 was significantly higher than HL-60 MF. The higher non-specific bindings of OCI-AML3 control were explained by the higher side scatter (SSC) values for OCI cells noted on bivariate dot plots. These findings are illustrated in Figure 1, showing single parameter histograms of HL60 and OCI-AML3 labeled with anti-pan NPM (Fig. 1a, b) and with wtNPM (Fig. 1c, d), and bivariate dot plots of HL60 and OCI-AML3 negative controls for the anti-pan test (Fig. 1e, f). The remaining four histograms represent the negative control for each test, where Figure 1g and 1h are the HL60 and OCI-AML3 negative controls for the anti-pan test, while Figure 1i and 1j are controls for the wtNPM test.
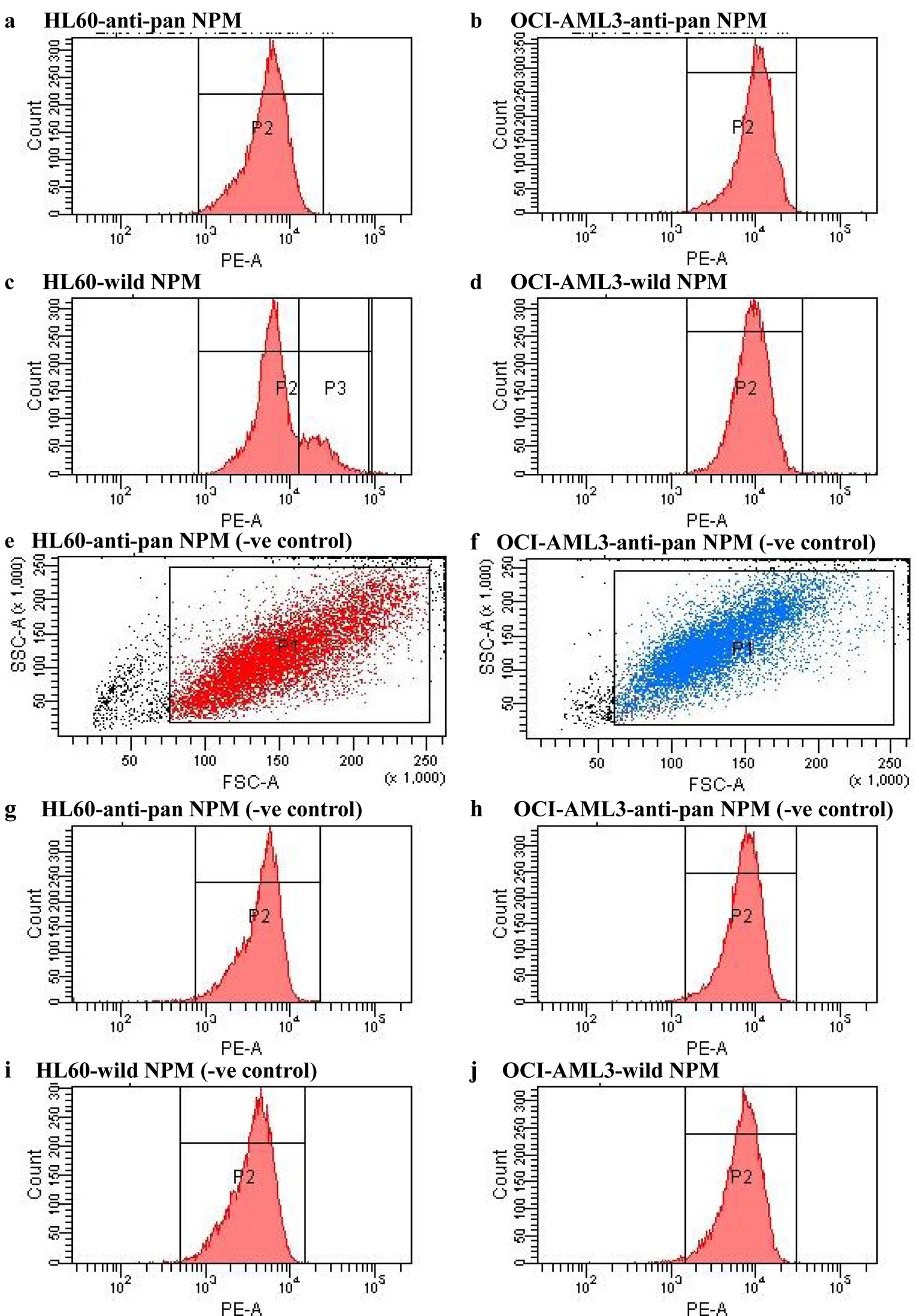 Click for large image | Figure 1. FACS analysis of HL60 and OCI-AML3 cells labeled with specific anti-NPM antibodies including anti-pan NPM and antibody for wild-type NPM, and their equivalent negative controls as labeled in the figure. NPM: nucleophosmin; FACS: fluorescence-activated cell sorting; AML: acute myeloid leukemia. |
Mutated NPM fluorescence was expressed as the ratio of the anti-pan NPM MF to wtNPM MF, for each cell type separately, by referring to Equation (1): the higher the ratio the more mNPM expressed by the analyzed cells.
It was apparent that wtNPM MF has a double peak in hl60, but not the OCI-AML3 or the HL60 control. Thus, HL60 wtNPM MF was calculated using the arithmetic mean, Equation (2).
Mathematical calculations were done using the mean fluorescence values obtained from the statistical view provided by the FACSDiva program, and summarized in Table 1. After calculations the ratio of anti-pan NPM MF/wt NPM MF for OCI-AMl3 was 1.6, while that for HL60 was 0.38.
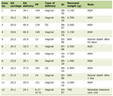 Click to view | Table 1. Numerical Values of Flow Cytometeric Analysis of Human Leukemia Cell Lines, HL60 and OCI-AML3 Labeled With wtNPM and Anti-Pan NPM, and Their Equivalent Negative Controls |
From the above mentioned straightforward mathematical analysis it is apparent that the ratio of anti-pan NPM fluorescence is higher for the OCI-AML3 compared with HL60. This difference between the two ratios together with what has been reported by Quentmeier et al that OCI-AML3 bears mNPM in their cytoplasm [16] provides evidence that FCM is a simple, available technique for quantitative assay of mNPM.
Differential expression of wtNPM within the same sample
To further confirm and explain the MF double peak of wtNPM in HL60, we carried out flow cytometeric analysis of HL60, and OCI-AML3 cells labeled with NPM FC61991 antibody, using the same method described before. However, we used a different absorption and emission wave lengths chromophobe, namely FITC; for data display and analysis we used the bivariate dot plots together with the single parameter histograms. FACS analysis revealed a mean fluorescence double peak in labeled HL60 cells, while both HL60 control and OCI-AML3 have a single peak MF.
The histogram illustrated in Figure 2 showed a double peak MF of wtNPM in HL60 cells. This double peak was mainly due to the presence of two populations, one was dim (P1) with a fluorescence intensity ranged between the third and fourth decades, while the other (P2) was bright with intensity ranged between the fourth and fifth decades. From this histogram and the statistical view summarized in Table 2, it is clear that the dim population is higher in number, but has lower MF values of 17,282, while the bright one is smaller in number with higher MF of 28,436.
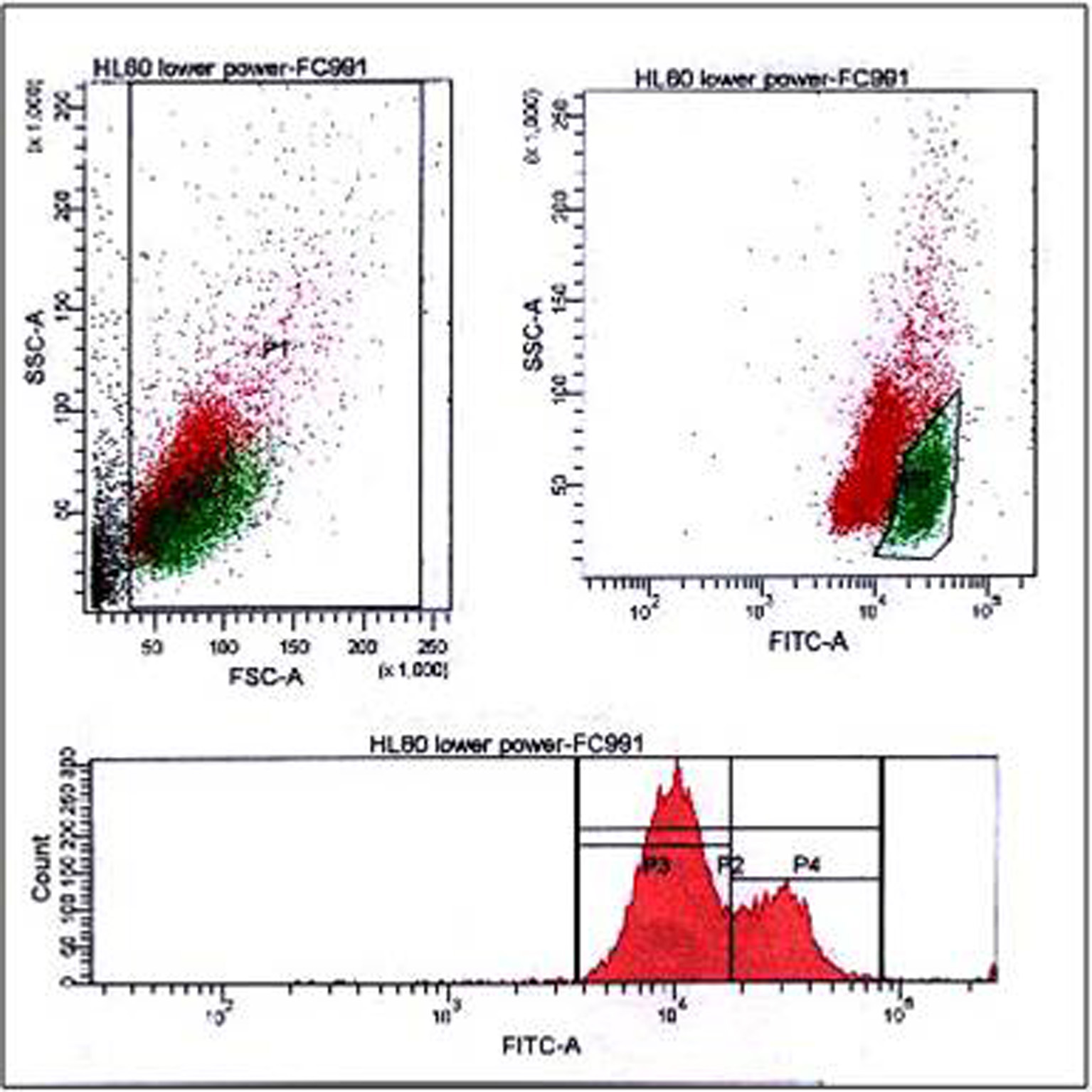 Click for large image | Figure 2. FACS analysis of HL60 cells labeled with wtNPM, showing dual populations in the bivariate dot plots (upper panel), quiescent (red) and dividing (green), both cellular debris and dividing cells were gated as indicated in the figure, while the lower panel showing a humped peak single parameter histogram. wtNPM: wild-type nucleophosmin; FACS: fluorescence-activated cell sorting. |
 Click to view | Table 2. FACS Data of HL60 and OCI-AML3 Labeled With wtNPM, and HL60 Negative Control |
Results presented and analyzed by scatter gram using SSC versus forward scatter (FSC) and examining the associated fluorochrome expression showed that this double peak is due to presence of these two cellular populations with the smaller population bigger in size and expressing more wtNPM. On the contrary both HL60 negative control and labeled OCI-AML3 had a single peak MF histogram; furthermore, this single peak was represented with a single population of cells in the scatter gram (Figs. 3, 4).
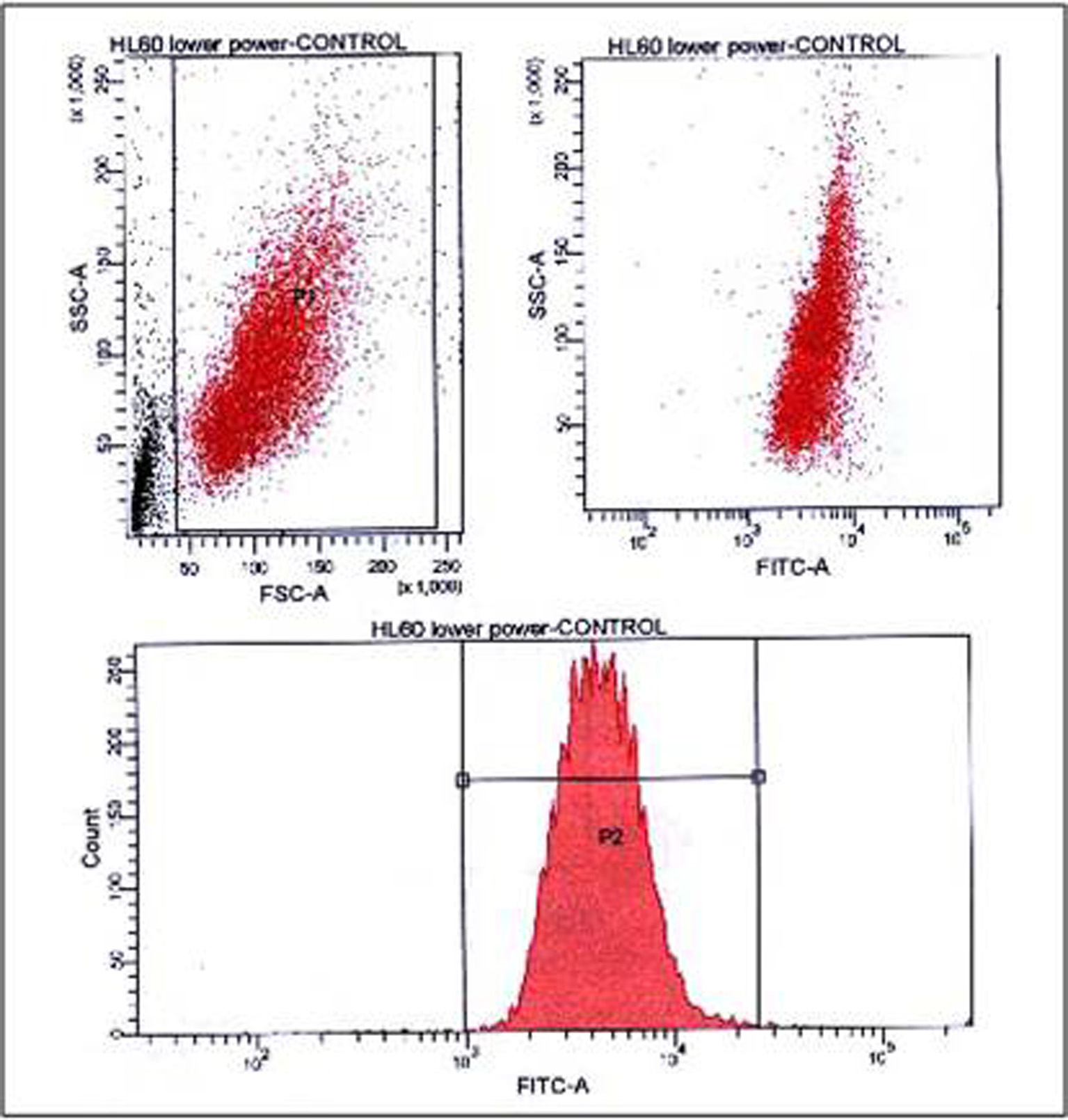 Click for large image | Figure 3. FACS analysis of HL60 negative control (secondary antibody only), showing presence of single population in the bivariate dot plots (upper panel), debris was gated as indicated, and a single peak MF histogram in the lower panel. FACS: fluorescence-activated cell sorting. |
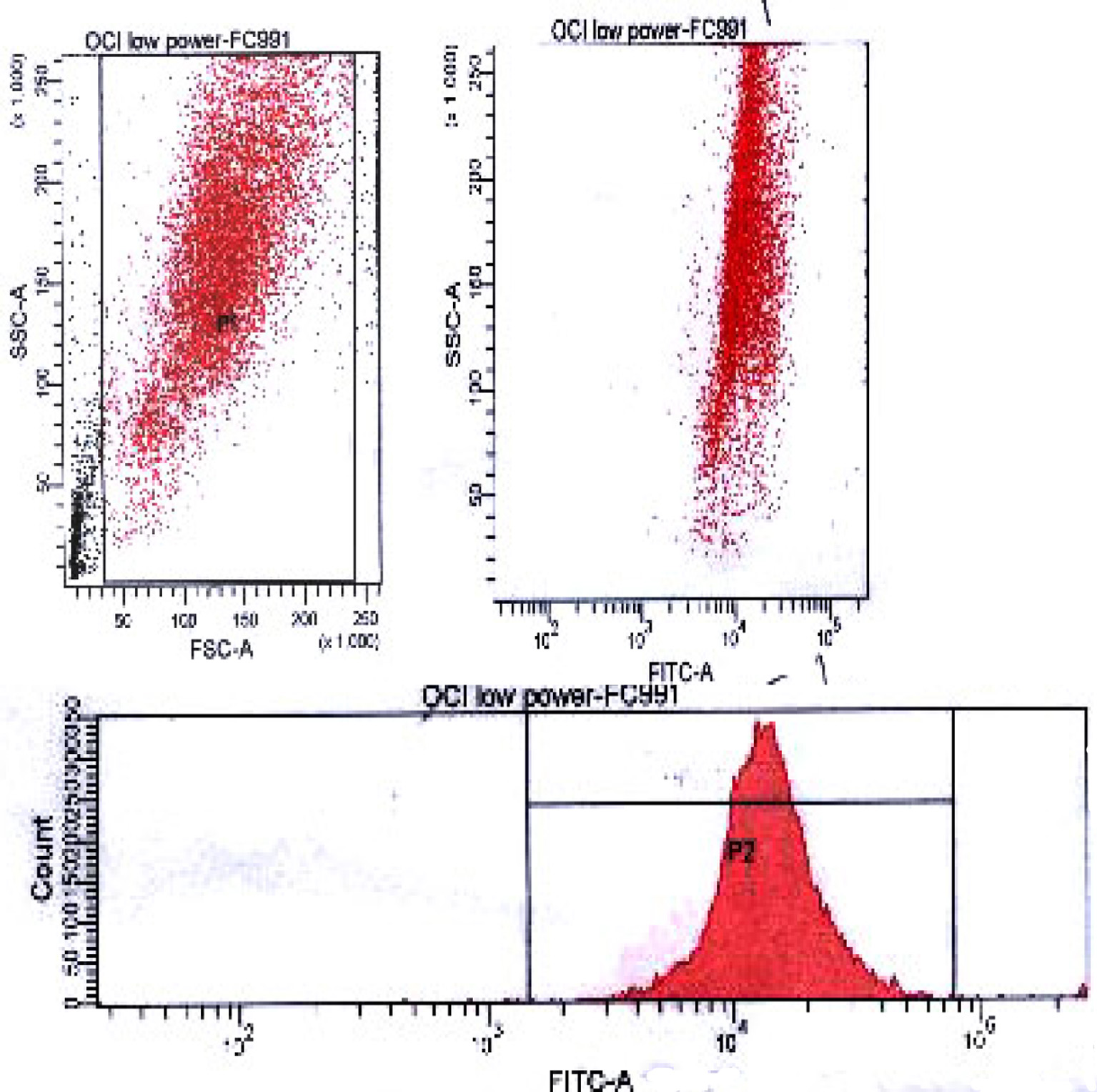 Click for large image | Figure 4. FACS analysis of wtNPM in OCI-AML3 cells, showing a single population in the bivariate dot plots (upper panel), debris was gated as indicated, and a single peak MF histogram in the lower panel. FACS: fluorescence-activated cell sorting; wtNPM: wild-type nucleophosmin; AML: acute myeloid leukemia. |
| Discussion | ▴Top |
These findings could not be explained that the dim ones are dead cells or cellular debris, as FCM has the ability to differentiate dead cells from living ones by differences in their scatter properties [17]. Dead cells have lower FSC and higher side scatter than living cells. Although fixation is an important step in our method and that all cells after fixation are dead, this important criterion is still preserved. The most reasonable explanation is the presence of two different populations in the same sample; one of them is mostly dividing and has a bigger size and more abundant wtNPM, while the other seems to be quiescent, smaller in size and expresses much less wtNPM. However, this was not the case with OCI cells. This could be further explained by our previous detection of difference in the exponential growth rate between the two cell types [16], where the estimated doubling time for HL60 is approximately 12 - 24 h, while that of OCI is 18 - 24 h.
To further investigate these two populations, cells were extracted from the same HL60 flask, and two cytospins were prepared. One was immunostained with wtNPM, while the other was incubated with TR secondary antibody only (control). They were analyzed with the immunofluorescence microscope. The obtained immunostaining pattern revealed presence of two groups of cells, resting cells with strict nucleolar localization of wtNPM, and dividing ones with nucleocytoplasmic localization. An interesting finding was that there was no staining detected in the control cytospin which ensures absence of non-specific bindings, as shown in Figure 5. These findings further explained the humped peak MF of wtPM in HL60 cells. An interesting finding was the typical nucleolar localization of wtNPM in resting cells, where NPM was clearly sparing the nucleolar central region. This finding reaffirmed our previous finding that the nucleolar fibrillar region is occupied with the fibrillarin rather than NPM [18].
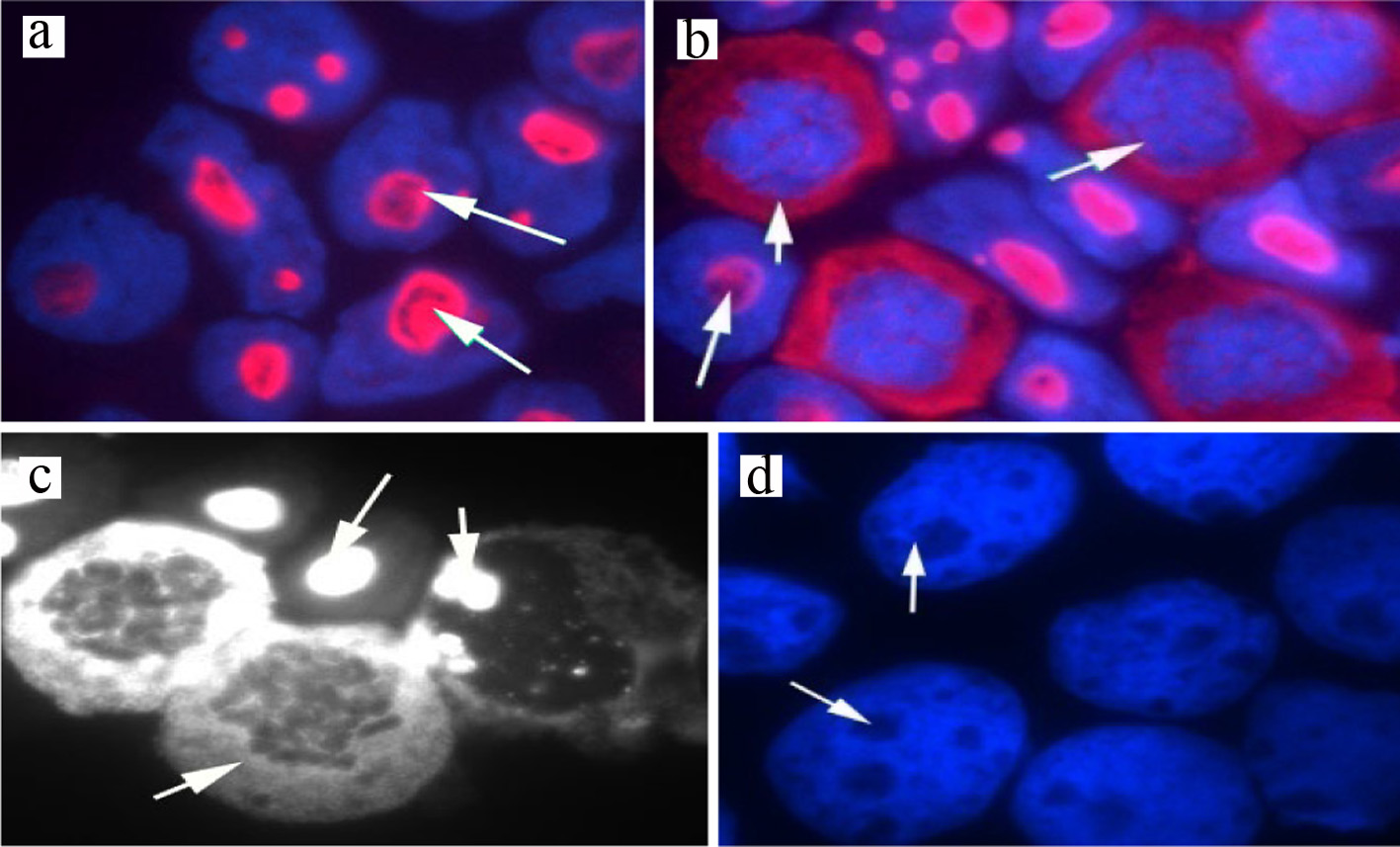 Click for large image | Figure 5. Immunostaining of wtNPM in (a) resting HL60 showing nucleolar localization (red arrowed), (b) dividing HL60 showing cytoplasmic localization in the arrowed cells, images were captured with the Hamamatasu camera and the nuclei were stained with DAPI (blue), (c) HL60 cells stained with wtNPM (white) image, grabbed with the Nikon Eclipse black and white camera, (d) HL60 negative control stained with Texas Red-conjugated secondary antibody only (no staining detected). wtNPM: wild-type nucleophosmin; DAPI: 4',6-diamidino-2-phenylindole. |
Advantages of using FCM in measuring the cellular protein NPM over PCR assay are dependent mainly on its wide availability and technical simplicity. Also, FCM has the ability for straightforward quantification of the amount of the assayed NPM, rather than extracting it from the amount of PCR product, and thus yields more precise and accurate results. Moreover, FCM is an intelligent machine that identifies dying cells and cellular debris, and allows their extraction from the analysis through the gating facility. This highly important facility is not available at the PCR method of assay, and the dead cells are capable of producing positive PCR signals, which adds to the inaccuracy and the possibility to have false positive results [10].
On the other hand, peripheral blood samples, or bone marrow aspirates are the best materials for flow cytometric assay of NPM mutational analysis, which was attributed to their fluid nature. These materials are mandatory to be obtained in each individual patient of AML, not only to confirm diagnosis but also to help follow-up and disease monitoring thereafter [19-21]. This important feature adds to the feasibility of FCM in measuring mNPM.
Conclusions
Conclusively, we developed a simple assay of mNPM with FCM. Furthermore, we demonstrated that the multiparameter FCM has the ability to deliver precise quantitative measurement of mNPM, although the value of quantification was not clear. However, the study illustrated that FCM is a highly important tool in AML prognosis as well as in AML diagnosis, and that flow cytometric detection of mNPM is a reliable technique which can be recommended for further substratification of the intermediate risk AML group into various prognostic subgroups.
Next, we carried the investigation further and analyzed the presence of two populations in one sample. The analysis revealed that NPM is expressed differently in each population. This finding was further confirmed with the results of immunostaining of NPM in cells from the same sample, which showed that NPM is nucleolarly expressed in one group of cells while cytoplasmically localized in the other. These findings provided evidence that FCM is not only a quantitative mean for comparing NPM expression between different patients, but it can also differentiate NPM expression among cells from the same patient. This criterion adds to the accuracy of the flow cytometeric method in measuring NPM and the potential to use this method for minimal residual disease (MRD) monitoring.
The validity of our screening method of mutated NPM in molecular grouping of AML patients was strengthened by the recent introduction of mNPM in the WHO classification of myeloid neoplasms. That now incorporates AML with mutated NPM (synonym NPMc+) as a new entity [22].
| Recommendations | ▴Top |
In view of the above mentioned findings, we hypothesized that mNPM analysis with FCM should be an integral part of primary AML risk stratification. Also, we recommended that this step should be combined with or second to immunophenotyping in the diagnostic/prognostic workup of AML. This is based on results of previous studies, which revealed that the concurrent presence of mNPM and other cytogenetic abnormalities have not been reported in the same patient or other myeloid neoplasms [6, 23]. Thus, the diagnostic/prognostic hierarchy should include NPM mutational analysis before the cytogenetic analysis. Accordingly, patients with NPM gene mutation will be considered to have normal karyotype and allocated in the intermediate risk group. Those with wtNPM have one of two possibilities either to be bearing different chromosomal abnormalities or to be of normal karyotype but did not express mNPM. In this later group, further cytogenetic studies are recommended.
Moreover, we assumed that the flow cytometeric assay of mutated NPM is the most suitable tool for minimal disease monitoring in AML patients bearing NPM gene mutation. This assumption was based on the observation of the stable nature of NPM mutation over the course of the disease, where it was reported that patients with NPM mutation at diagnosis lost the mutation at complete remission, and acquire the same mutation at relapse. On the other hand, patients without NPM mutation at diagnosis did not acquire NPM mutation neither during follow-up nor at relapse [8, 24-26].
The key issue behind using mNPM for MRD monitoring is the detection of the precise type of NPM mutation in each individual patient. In this context FCM has the potential to deliver accurate measurement of each single NPM moiety specifically. However, flow cytometeric specificity is dependent mainly on the specificity of the antibody used in the assay, thus the validity of mNPM in MRD monitoring will be better with improving specificity of the commercially available anti-NPM antibodies to be targeting single NPM mutational type [27-29].
Future prospective
After development of accurate quantitative assay of mNPM in this research, further large scale clinical studies of this assay are still needed. In these studies the obtained mNPM levels should be thoroughly followed. The main aim is to correlate mNPM over- or under-expression with various disease outcomes and allow its use to stratify patients with normal karyotype into further prognostic subgroups.
Further research is needed to improve and broaden the panel of the commercially available anti-human NPM antibodies, especially those targeting the mutated type, to distinguish the precise type of mutation. Accordingly, flow cytometeric assay of mNPM could be applied for MRD monitoring in patients with AML.
Acknowledgments
We would extend our thanks to Julie Adams at Clinical Hematology Department, Manchester Royal Infirmary, UK, for her expert guidelines, and also to Dr. Karen-Rees Unwin and Dr. Suzanne Johnson, at Manchester School of Medicine, UK, for their guidance throughout the work. We greatly thank the Egyptian Cultural Bureau in London for the great support they offered during this work.
Financial Disclosure
This work was supported by the Egyptian Cultural Bureau educational grant on behalf of the Ministry of Higher Education.
Conflict of Interest
There was no known conflict of interest associated with this work.
Informed Consent
Not applicable.
Author Contributions
JB suggested the research point, formulated objectives and put forward the research plan. SAAK is the principal investigator, who did the research work, interpreted the obtained results together with JB, and edited the paper. EBEAE, LFE, HMA and SMA helped in getting the grant, provided support for the investigators during the work, and revised the work meticulously.
| References | ▴Top |
- Hou HA, Lin CC, Chou WC, Liu CY, Chen CY, Tang JL, Lai YJ, et al. Integration of cytogenetic and molecular alterations in risk stratification of 318 patients with de novo non-M3 acute myeloid leukemia. Leukemia. 2014;28(1):50-58.
doi pubmed - Schlenk RF, Dohner H. Genomic applications in the clinic: use in treatment paradigm of acute myeloid leukemia. Hematology Am Soc Hematol Educ Program. 2013;2013:324-330.
doi pubmed - Mrozek K, Marcucci G, Nicolet D, Maharry KS, Becker H, Whitman SP, Metzeler KH, et al. Prognostic significance of the European LeukemiaNet standardized system for reporting cytogenetic and molecular alterations in adults with acute myeloid leukemia. J Clin Oncol. 2012;30(36):4515-4523.
doi pubmed - Jabbour E, Kantarjian H. Chronic myeloid leukemia: 2012 update on diagnosis, monitoring, and management. Am J Hematol. 2012;87(11):1037-1045.
doi pubmed - Alyamany A, Khaled SAA, Farghaly R, Zedan A. Response to front and second line treatment in patients with acute non lymphoblastic leukemia: a single center experience. Annals of Hematology. 2019;98:S44-S44
- Falini B, Maria Paola Martelli, Niccolo Bolli, Rossella Bonasso, et al. Immunohistochemistry predicts nucleophosmin mutations in acute myeloid leukaemia. Blood. 2006;108:1999-2005.
doi - Falini B, Sportoletti P, Martelli MP. Acute myeloid leukemia with mutated NPM1: diagnosis, prognosis and therapeutic perspectives. Curr Opin Oncol. 2009;21(6):573-581.
doi pubmed - Chopra A, Soni S, Pati H, Kumar D, Diwedi R, Verma D, Vishwakama G, et al. Nucleophosmin mutation analysis in acute myeloid leukaemia: Immunohistochemistry as a surrogate for molecular techniques. Indian J Med Res. 2016;143(6):763-768.
doi pubmed - Ammatuna E, Noguera NI, Zangrilli D, Curzi P, Panetta P, Bencivenga P, Amadori S, et al. Rapid detection of nucleophosmin (NPM1) mutations in acute myeloid leukemia by denaturing HPLC. Clin Chem. 2005;51(11):2165-2167.
doi pubmed - Calvo KL, Ojeda MJ, Ammatuna E, Lavorgna S, Ottone T, Targovnik HM, Lo-Coco F, et al. Detection of the nucleophosmin gene mutations in acute myelogenous leukemia through RT-PCR and polyacrylamide gel electrophoresis. Eur J Haematol. 2009;82(1):69-72.
doi pubmed - Steinberg TH, Agnew BJ, Gee KR, Leung WY, Goodman T, Schulenberg B, Hendrickson J, et al. Global quantitative phosphoprotein analysis using Multiplexed Proteomics technology. Proteomics. 2003;3(7):1128-1144.
doi pubmed - Scholl S, Mugge LO, Landt O, Loncarevic IF, Kunert C, Clement JH, Hoffken K. Rapid screening and sensitive detection of NPM1 (nucleophosmin) exon 12 mutations in acute myeloid leukaemia. Leuk Res. 2007;31(9):1205-1211.
doi pubmed - Grimwade L, Gudgin E, Bloxham D, Bottley G, Vassiliou G, Huntly B, Scott MA, et al. Detection of cytoplasmic nucleophosmin expression by imaging flow cytometry. Cytometry A. 2012;81(10):896-900.
doi pubmed - Du Pisani LA, Shires K. Development of a flow cytometric method to detect the presence of mutated nucleophosmin 1 in acute myeloid leukemia. Hematol Oncol Stem Cell Ther. 2015;8(3):106-114.
doi pubmed - Herzenberg LA, Tung J, Moore WA, Herzenberg LA, Parks DR. Interpreting flow cytometry data: a guide for the perplexed. Nat Immunol. 2006;7(7):681-685.
doi pubmed - Quentmeier H, Martelli MP, Dirks WG, Bolli N, Liso A, Macleod RA, Nicoletti I, et al. Cell line OCI/AML3 bears exon-12 NPM gene mutation-A and cytoplasmic expression of nucleophosmin. Leukemia. 2005;19(10):1760-1767.
doi pubmed - Dalal BI. Clinical applications of molecular haematology: flow cytometry in leukaemias and myelodysplastic syndromes. J Assoc Physicians India. 2007;55:571-573.
- Khaled SA, Burthem J, Abo ElNoor E, et al. Role of nucleophosmin gene mutation in leukemogenesis of acute myeloid leukemia. J Hematol. 2018;7(1):7-13.
doi - Khaled SA, Burthem J, Abo El Noor E, El Toni L, Ahmed S, Ahmed H. Differences between nucleophosmin isoforms in de-novo acute myeloid leukemia: possible implications in developing targeted therapy for acute myeloid leukemia with normal karyotype. Egyptian J Hematol. 2015;40(41):190-194.
doi - Chou WC, Tang JL, Liang IL, et al. Nucleophosmin mutations in de novo acute myeloid leukaemia: the age dependent incidences and the stability during disease evolution. Cancer Res. 2006;66:3310-3315.
doi pubmed - Kebriaei P, de Lima M, Estey EH, Champlin R. Chapter 107: Management of Acute Leukemias. In: DeVita VT, Lawrence TS, Rosenberg SA, eds. DeVita, Hellman, and Rosenberg's Cancer: Principles and Practice of Oncology. 10th ed. Philadelphia, Pa: Lippincott Williams & Wilkins; 2015.
- Appelbaum FR. Chapter 98: Acute leukemias in adults. In: Niederhuber JE, Armitage JO, Dorshow JH, Kastan MB, Tepper JE, eds. Abeloff's Clinical Oncology. 5th ed. Philadelphia, Pa. Elsevier: 2014.
- Vardiman JW, Thiele J, Arber DA, Brunning RD, Borowitz MJ, Porwit A, Harris NL, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood. 2009;114(5):937-951.
doi pubmed - Chou WC, Tang JL, Lin LI, Yao M, Tsay W, Chen CY, Wu SJ, et al. Nucleophosmin mutations in de novo acute myeloid leukemia: the age-dependent incidences and the stability during disease evolution. Cancer Res. 2006;66(6):3310-3316.
doi pubmed - Caudill JS, Sternberg AJ, Li CY, Tefferi A, Lasho TL, Steensma DP. C-terminal nucleophosmin mutations are uncommon in chronic myeloid disorders. Br J Haematol. 2006;133(6):638-641.
doi pubmed - Suzuki T, Kiyoi H, Ozeki K, Tomita A, Yamaji S, Suzuki R, Kodera Y, et al. Clinical characteristics and prognostic implications of NPM1 mutations in acute myeloid leukemia. Blood. 2005;106(8):2854-2861.
doi pubmed - Bolli N, Galimberti S, Martelli MP, Tabarrini A, Roti G, Mecucci C, Martelli MF, et al. Cytoplasmic nucleophosmin in myeloid sarcoma occurring 20 years after diagnosis of acute myeloid leukaemia. Lancet Oncol. 2006;7(4):350-352.
doi - Schnittger S, Schoch C, Kern W, Mecucci C, Tschulik C, Martelli MF, Haferlach T, et al. Nucleophosmin gene mutations are predictors of favorable prognosis in acute myelogenous leukemia with a normal karyotype. Blood. 2005;106(12):3733-3739.
doi pubmed - Oelschlaegel U, Koch S, Mohr B, Schaich M, Falini B, Ehninger G, Thiede C. Rapid flow cytometric detection of aberrant cytoplasmic localization of nucleophosmin (NPMc) indicating mutant NPM1 gene in acute myeloid leukemia. Leukemia. 2010;24(10):1813-1816.
doi pubmed
This article is distributed under the terms of the Creative Commons Attribution Non-Commercial 4.0 International License, which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Journal of Hematology is published by Elmer Press Inc.


