| Journal of Hematology, ISSN 1927-1212 print, 1927-1220 online, Open Access |
| Article copyright, the authors; Journal compilation copyright, J Hematol and Elmer Press Inc |
| Journal website http://www.thejh.org |
Original Article
Volume 3, Number 2, June 2014, pages 34-42
Study of Correlation Between SFRP-1 and SFRP-2 Hypermethylation With Relapse, Complete Remission, Genetic Mutations of FLT3-ITD and NPM1 and Immunophenotypes of Leukemic Cells in Patients With De Novo Acute Myeloblastic Leukemia
Ali Ghasemia, Fatemeh Nadalia, Bahram Chahardoulib, Nasrin Alizad Ghandforosha, Ardeshir Ghavam Zadehb, Shahrbano Rostamib, c
aDepartment of Hematology, School of Allied Medical Sciences, Tehran University of Medical Sciences, Tehran, Iran
bHematology-Oncology and Stem Cell Transplantation Research Center, Tehran University of Medical Sciences, Tehran, Iran
cCorresponding Author: Shahrbano Rostami, Hematology-Oncology and Stem Cell Transplantation Research Center, Tehran University of Medical Sciences, Tehran, Iran
Manuscript accepted for publication May 27, 2014
Short title: SFRP-1 and SFRP-2 Hypermethylation
doi: https://doi.org/10.14740/jh154w
| Abstract | ▴Top |
Background: Acute myeloid leukemia (AML) is a clonal disorder of hematopoietic stem cells that is associated with the infiltration of blasts in the peripheral blood and bone marrow. In recent years, the role of epigenetic abnormalities such as promoter methylation of tumor suppressor genes, including secreted frizzled related proteins (SFRPs) family genes, has been shown in the pathogenesis of cancers. This study evaluates the correlation between SFRP-1 and SFRP-2 hypermethylation with genetic mutations of FLT3-ITD, NPM1 and immunophenotypes of leukemic cells.
Methods: In peripheral blood from 43 patients with de novo AML, isolated DNA was treated with sodium bisulphite and analyzed by methylation-specific polymerase chain reaction (MSP). Fisher exact test and SPSS 21 were used for statistical data analysis.
Results: SFRP-1 hypermethylation had significant association with CD34 (P = 0/045) and CD14 (P = 0/046) expression. There was no difference in the incidence of genetic mutations of FLT3-ITD and NPM1 between patients with and without SFRP hypermethylation.
Conclusion: Based on these results, hypermethylation of SFRP-1 is associated with increased expression of CD34 and CD14 antigenes. However, further studies on a large group of patients are necessary to confirm our findings.
Keywords: Acute myeloblastic leukemia; DNA methylation; Genetic mutation; Immunophenotype
| Introduction | ▴Top |
Acute myeloid leukemia (AML) is a clonal hematopoietic disorder characterized by uncontrolled self-renewal of hematopoietic stem cells, maturation arrest at myeloblast level, peripheral blood and bone marrow infiltration of blast cells [1]. It is demonstrated that pathogenesis of AML is associated with some disorders including genetic changes and chromosomal translocations. Developments in molecular researches have improved our understanding of the leukemogenesis in AML. In addition to age, white blood cells count and cytogenetic aberrations, investigations of molecular genetic alterations affecting NPM1 (nucleophosmin1), FLT3 genes and WT1 (Wilms’ tumor) assay are known as important prognostic factors in AML. In recent years, epigenetic disorders including methylation of tumor suppressor genes like secreted frizzled related proteins (SFRPs) family genes have also been shown to play a role in AML pathogenesis. These alterations may lead to differentiation and apoptosis arrest in leukemic blasts as well as increase in proliferation and self-renewal [2]. SFRPs are Wnt antagonists that suppress this signaling pathway in healthy individuals. Wnt signaling pathway contributes to regulation of cell proliferation and differentiation. In some malignancies like colorectal cancers, head and neck tumors and gastric cancer, aberrant Wnt signaling pathway has been shown to cause uncontrolled cell proliferation [3]. Chronic myeloid leukemia was the first malignancy in which the important role of Wnt signaling pathway has been described [4]. β-catenin is an intracellular regulator of transcription that is associated with cancers. Wnt controls the cytoplasmic level and stability of β-catenin [5]. In absence of Wnt ligand and its protectional role, β-catenin level decreases due to destruction by casein kinase 1 and glycogen synthase kinase 3b enzymes [6]. But when the ligand adheres to its receptor (frizzled receptor), it activates DV1 (dishevelled) proteins [7]. Having accumulated in cytoplasm, β-catenin migrates to nucleus where it causes expression of some genes involved in cell proliferation and differentiation [3, 8]. It has recently been demonstrated that both chromosomal alterations and FLT-3 mutations associated with AML pathogenesis affect Wnt signaling pathway [9]. Methylation of SFRP gene leads to loss of its inhibitory effect on Wnt pathway. Then cytoplasmic and nuclear levels of β-catenin enhances that as a transcription factor makes some genes associated in cell cycle regulation like MYC and cyclin D to be expressed [10]. Since the methylation of these genes may play a role in initiation and leukemogenesis of AML, in present study, we investigated the methylation status of SFRP-1 and SFRP-2 genes in newly diagnosed AML patients and correlation between SFRP-1 and SFRP-2 hypermethylation with clinical and laboratory findings such as relapse, complete remission, genetic mutations of FLT3-ITD and NPM1 and immunophenotypes of leukemic cells that have been accepted in Hematology, Oncology and Bone Marrow Transplantation Research Center of Tehrans Shariati Hospital.
| Materials and Methods | ▴Top |
Patients
Peripheral blood samples were drawn from 25 healthy individuals as negative control group and from 43 patients with newly diagnosed AML that were accepted in Hematology, Oncology and Stem Cell Transplantation Research Center, Shariati Hospital, Tehran, Iran. All patients were classified according to morphologic and immunophenotypic (FAB) criteria. A panel of mono-clonal antibodies designed against myeloid lineage specific antigenes including CD33, CD11b, CD15, CD14, CD64, CD117 and MPO and non-specific antigenes including CD34 and HLA-DR based on previous studies done [11, 12].
DNA extraction and bisulfite treatment
Mononuclear cells of drawn samples including leukemic blast cells were isolated by concentration gradient sedimentation using Ficoll-hypaque. Genomic DNA was extracted by saturated salt standard method [11]. In the next step, extracted DNA underwent bisulfite conversion with the Epitect Bisulfite kit (Qiagen) using producer instruction. By this treatment unmethylated cytosines converted to uracil where methylated cytosines stayed intact.
MSP methylation analysis
The methylation status of SFRP-1 and SFRP-2 genes was investigated using methylation-specific PCR (MSP) technique. In this method, we used two pairs of primers specified for checking the methylated or unmethylated residue. These primers are given in Table 1 accompanied with product values. The sequences of these primers are designed in previous studies [13, 14].
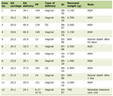 Click to view | Table 1. SFRP-1 and SFRP-2 Gene Primers Sequences, Annealing Temperature and Product Size for MSP Assays |
Four MSP reactions using methylated and unmethylated primers related to SFRP-1 and SFRP-2 were administered for each patient. In methylation testing, we used 2 µL of DNA previously treated with bisulfite, 4.5 µL of dH2O, 12.5 µL of master mix, 0.5 µL of forward primer and 0.5 µL of reverse primer while in order to investigate the unmethylated status, we used 2 µL of DNA, 8.5 µL of dH2O, 12.5 µL of master mix, 0.5 µL of forward primer, 0.5 µL of reverse primer and 1 µL of MgCl2. In the first step of MSP, reaction components were put in pre-thermal condition including 98 °C for 1 min and 96 °C for 3 min followed by 40 cycles including 99 °C for 10 s, 97 °C for 20 s, 54 °C for 30 s (SFRP-1, UM primer), 64 °C for 30 s (SFRP-1, 2-M primer) and 72 °C for 7 min (extension). In this study, we used EpiTect PCR control DNA kit (Qiagen Inc., cat no. 59695) containing unmethylated and completely methylated DNAs as negative and positive controls, respectively. ddH2O served as a blank control. Electrophoresis on 2.5% agarose gel was done for MSP product identification.
NPM1 and FLT3-ITD mutations detection by capillary electrophoresis
Primer sequences for detection of mutations in nucleophosmin (NPM1) exon 12 and FLT3-ITD exon 14 are given in Table 2. Forward primers for fragment length analysis of FLT3-ITD and NPM1 were fluorescently labeled with two different colors (FAM, ROX respectively) and mixed in multiplex PCR reactions. The PCR reaction contained 50 ng of genomic DNA, 0.3 µM of each primer, 12.5 µL of 2 × Taq PCR master mix (Qiagen) and 1.25 µL Q-solution (Qiagen). Samples were amplified using the following PCR conditions: 95 °C, 30 s for denaturation; 62 °C, 30 s for primer annealing; and 72 °C, 30 s for elongation, repeated for 35 cycles. Afterwards the PCR products were mixed with a size marker (GeneScan™ 500 LIZ™ Dye Size Standard; Applied Biosystems, Foster City, CA, USA) and then electrophoresed on an ABI 3130 Genetic Analyzer (Applied Biosystems) and the results were analyzed using the GeneMapper v3.5 software (Applied Biosystems).
 Click to view | Table 2. Primer Sequences for Detection of Mutations in Nucleophosmin 1 (NPM1) Exon 12 and FLT3-ITD Exon 14 |
Statistical analysis
Statistical Package for the Social Sciences (SPSS) v 21 was used for statistical analysis. Pairwise comparisons between patients characteristics were performed by Fisher’s exact test for categorical variables and independent t-test for continuous variables. Mann-Whitney U-tests were used to compare continuous variables and medians of distributions. P value less than 0.05 were considered significant statistically.
| Results | ▴Top |
Of the 43 patients studied, 31 patients were male (72/1%) and 12 were female (27/9%). The age range between 15 and 72 years and a mean age of 4/45 years. SFRP-1 gene found hemi-methylated in 13 patients (30.2%), completely methylated in 13 patients (30.2%) and completely unmethylated in 17 patients (39.5%) (Fig. 1), while SFRP-2 gene was hemi-methylated in 16 patients (37.2%), completely methylated in nine patients (20.9%) and completely unmethylated in 18 patients (41.8%) (Fig. 2). None of control individuals showed methylation in SFRP-1 and SFRP-2 genes. Correlations between hypermethylation of SFRP-1 and -2 genes and relapse, complete remission and genetic mutations of FLT3-ITD and NPM1 are indicated in Table 3.
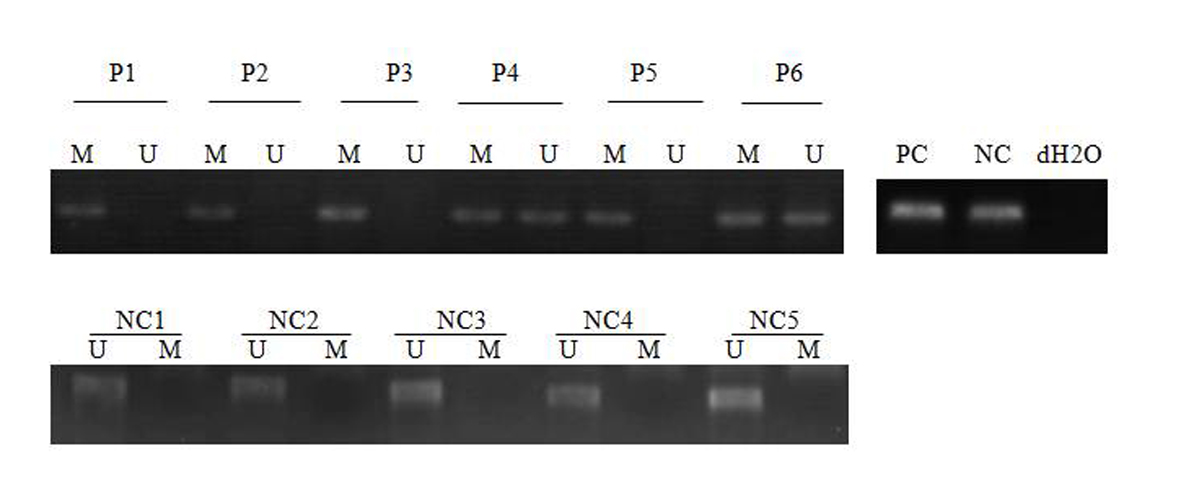 Click for large image | Figure 1. MSP analysis of SFRP-1 in AML patients and normal control. PC: positive control; NC: negative control; NC: normal control; P: patient; M: methylated; U: unmethylated. dH2O served as a blank control. |
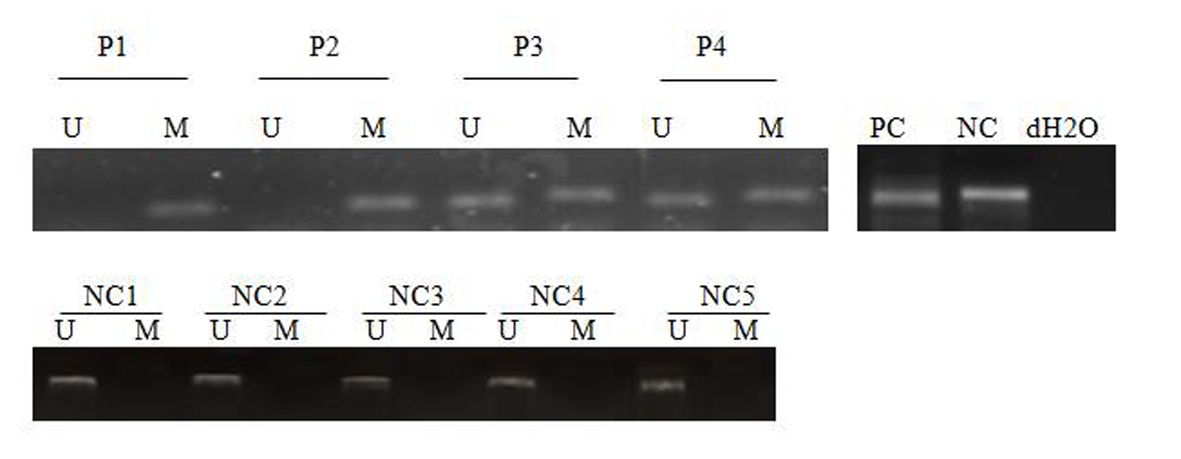 Click for large image | Figure 2. MSP analysis of SFRP-2 in AML patients and normal control. PC: positive control; NC: negative control; NC: normal control; P: patient; M: methylated; U: unmethylated. dH2O served as a blank control. |
 Click to view | Table 3. Correlations Between Hypermethylation of SFRP-1 and -2 Genes and Laboratory and Clinical Symptoms of AML Patients |
In AML patients, hypermethylation frequencies of SFRP-1 and SFRP-2 genes were 30.23% (13 out of 43 patients) and 20.9% (nine out of 43 patients), respectively. Also 32.2% of patients (13 out of 43 patients) showed methylated SFRP-1 and SFRP-2 genes at the time of diagnosis (Table 3). Aberrant methylations of these genes are found in all FAB classifications of AML except M6. Hypermethylations of SFRP-1 (P = 0.028) and SFRP-2 (P= 0.004) genes were associated with FAB-M0 subtype of AML (Table 2). There is no significant relationship between hypermethylation of SFRP-1 and SFRP-2 genes with clinical parameters of patients including sex, age, white cell and platelet (Table 3).
Six out of 43 subjects (4.6%) developed relapse, among whom two patients attributed to SFRP-1 gene and one patient (2.3%) for SFRP-2 gene were hypermethylated. There is no any significant relationship between hypermethylation of both SFRP-1 and SFRP-2 genes and relapse of patients. Also, information on the treatment of 38 patients (88.37%) was found. Of these number, 28 patients (75.67%) had complete remission after induction therapy, of whom nine and five patients were hypermethylated in the SFRP-1 and SFRP-2 genes, respectively. Ten patients (24.33%) were refractory to induction therapy, of whom three and two patients had hypermethylation in the SFRP-1 and SFRP-2 genes, respectively. There is no significant relationship between hypermethylation in the SFRP-1 and SFRP-2 genes among patients who developed whether methylation or not and complete remission after induction therapy (Table 3).
Capillary electrophoresis was used to discover the NPM1 and FLT3 mutations. NPM1-wt displayed consistently a single 236 bp peak and NPM1-mut bands were displayed at approximately 240 bp (Fig. 3). FLT3-wt was displayed at approximately 329 bp while the FLT3-ITD had two peaks 329 bp and another bigger, depending on the size of insertion (Fig. 3). FLT3-ITD and NPM1 mutations among 43 tested patients were found in eight (18.6%) and six patients (13.9%), respectively. From patients having SFRP-1 hypermethylation only two patients showed mutation in FLT3-ITD and only two patients (15.3%) had NPM1 mutation. Of SFRP-2 hypermethylation cases, three patients (33.3%) showed FLT3-ITD mutation also and two patients (22.2%) were demonstrated to have NPM1 mutation simultaneously. No significant association between SFRP-1 and SFRP-2 genes promoter hypermethylation and mutation of FLT3-ITD (PSFRP-1 = 0.999 and PSFRP-2 = 0.332 ) and NPM1 genes (PSFRP-1 = 0.999 and PSFRP-2 = 0.589) were found (Table 3). Between SFRP-1 hypermethylation and increased expression of CD34 (P = 0.045) and CD14 (P = 0.046), we found significant association. While no other antigene expression showed association with SFRP-1 hypermethylation, no significant associations were seen in immunophenotypic features of leukemic blast cells with or without SFRP-2 hypermethylation (Table 4).
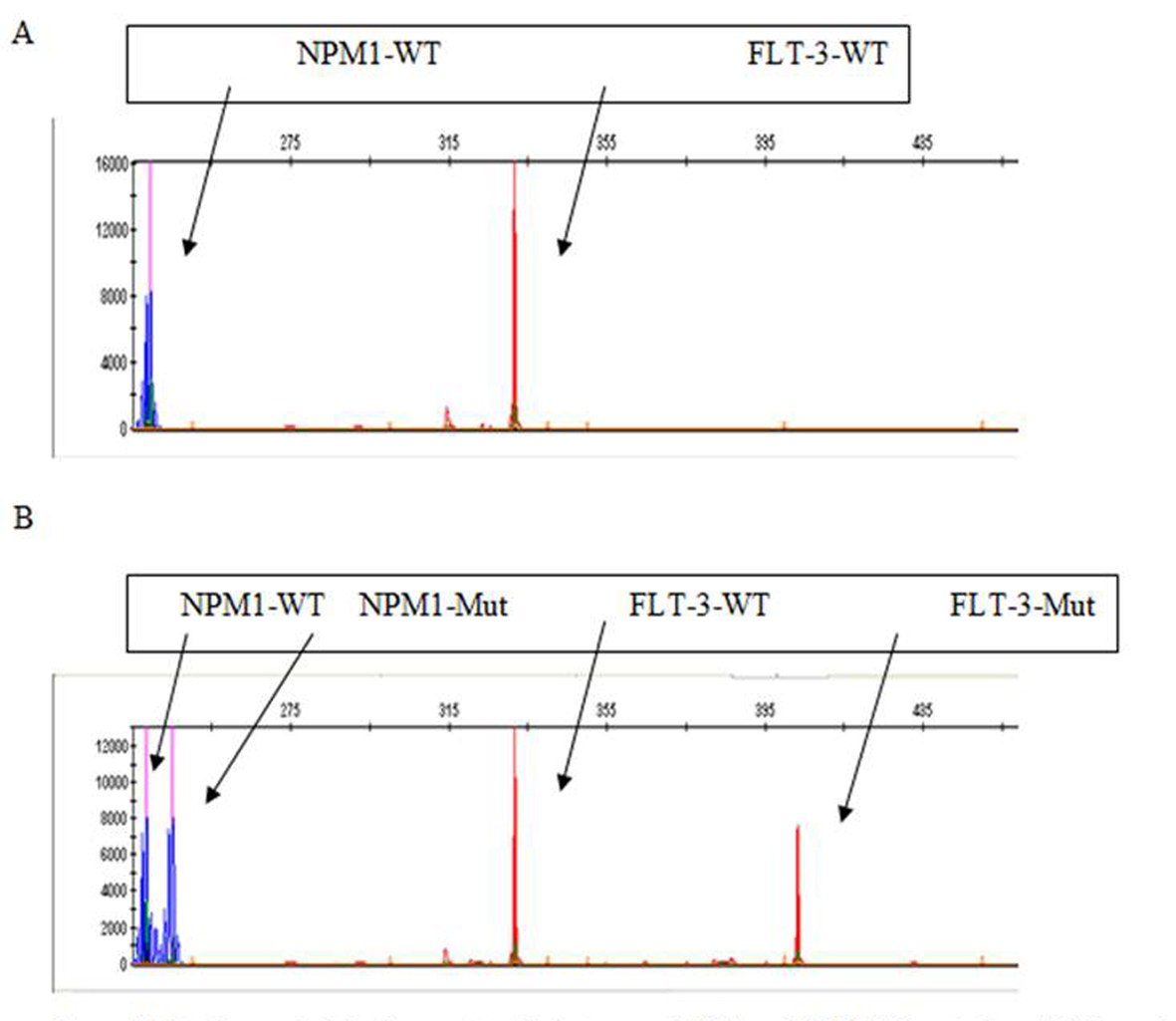 Click for large image | Figure 3. Mutation analysis by fragment analysis to assess NPM1 and FLT3/ITD mutations. (A) The peaks in blue and red show the results from amplification of the unmutated NPM1 and FLT3 alleles, respectively. (B) The peaks in blue and red on the right of the corresponding wild type alleles represent the mutated NPM1 and FLT3-ITD-positive alleles, respectively. |
| Discussion | ▴Top |
Wnt/β-catenin signaling pathway has been implicated in many cellular procedures including proliferation, morphology, motions, destiny determination of cells and organ development [15]. Understanding the roles of Wnt/β-catenin signaling in survival, proliferation and differentiation of hematopoietic stem cells resulted in developing the hypothesis that this signaling pathway may be involved in leukemogenesis [11, 15, 16]. SFRP is a tumor suppressor protein that modulates the Wnt/β-catenin signaling pathway. This protein binds to Wnt protein and thus inhibits its binding to Wnt-frizzled receptor and the result is inactivation of Wnt signaling pathway. Hence there may be an association between methylation of Wnt signaling antagonist genes and the activation of this pathway in leukemia and solid tumors [11, 12, 16]. Aberrant methylation of tumor suppressor genes is a more specific and common genetic event in human cancers [17, 18]. In present study, we investigated the methylation status of SFRP-1 and SFRP-2 genes promoters in 43 AML patients at the time of diagnosis and its correlation with relapse, complete remission, mutations of FLT3 and NPM1 genes as well as with immunophenotypic features of leukemic blast cells. The results of this study showed that hypermethylation of SFRP-1 and SFRP-2 genes occurs with a frequency of 30.23% (13 out of 43 subjects) and 20.9% (nine out of 43 patients) in AML patients at the time of diagnosis, respectively. While none of the normal samples showed methylation, Veeck et al demonstrated that epigenetic changes of SFRP-1 through methylation even linked to poor prognosis in patients with breast cancer [19, 20]. Cooper et al suggested that recombinant SFRP-1 may be a novel therapeutic strategy for cancers with suppressed SFRP-1 expression [21]. Moreover SFRP-2 methylation as one of the epigenetic targets occurs in cancers such as colon cancer [22], esophagus cancer [14], bladder cancer [9], gastric cancer [23, 24], liver cancer [25] and lung cancer [13]. Like previous studies, our study also showed that SFRP-1 and -2 genes are epigenetic modulation targets in AML patients which inactivated following methylation; therefore, methylation of these genes may be involved in the onset of AML. Indeed, because of the 43 patients studied, 26 patients (60.5%) for SFRP-1 gene and 25 patients (58.2%) for SFRP-2 gene had at least one methylated allele. Hemi-methylated patients may develop over time and it is far poor prognosis than do their occurrence. Methylation of SFRP genes has also been shown in hematologic malignancies, as Pehlivan et al showed that the activation of Wnt signaling pathway through methylation of SFRP-1 leads to drug resistance in patients with chronic myeloid leukemia treated with imatinib mesylate by suppression of imatinib mesylate effect over BCR-ABL signaling pathway [26]. Wang et al showed that methylation of SPRP genes in patients with MDS is associated with poor prognosis and less survivability [12]. In our study, we did not observe any significant association between hypermethylation of these genes and conventional prognostic factors in AML, like age and WBC count. Also no significant relationship was observed between methylation of these genes and other clinical parameters like sex, platelet count and hemoglobin. While that may be associated to be seen with an increased sample size, Hou et al showed that patients with FAB M0 subtype of AML had the highest incidence of hypermethylation of Wnt inhibitors, whereas those with M4/M5 subtype had the lowest incidence [27]. Our results also showed that aberrant methylation of these genes occurs in all the study FAB-AML subgroups including M0, M1, M2, M4 and M5, except subgroup M6. While the highest incidence of hypermethylation of SFRP-1 (P = 0.28) and SFRP-2 (P = 0.004) in M0 subgroup compared with the lowest incidence SFRP-1 (P = 0.91) and SFRP-2 (P = 0.936) takes place in M1 and M4 subgroups, respectively. Complete remission after induction therapy was observed in 60-80% of patients. In the present study, no significant association was observed between hypermethylation of SFRP-1 and SFRP-2 and complete remission after induction therapy and the response to treatment was identical in patients with and without hypermethylation. It has recently been demonstrated that in AML patients, the mutation of FLT3 gene and chromosomal aberrations are associated with modulations in Wnt signaling pathway. Mizuki and Tickenbrock showed that FLT3 mutation in myeloid progenitors by frizzled-4 (one of the Wnt receptors) induction causes elevation of β-catenin level and thus Wnt signaling aberration [28, 29]. Based on these findings, FLT3 mutation may play a relative role in leukemogenesis by activating Wnt signaling pathway. No significant association was found between methylation of these genes and FLT3 mutation. No significant correlation was observed between SFRP-1 and -2 hypermethylation and NPM1 mutation in our study but it may be possible to find a significant association between them by using more samples, because Hou et al, by using more samples (269 patients) demonstrated that theirs is a significant association between SFRP hypermethylation and mutation in NPM1 and CEBPA genes [27]. We also used flowcytometry to determine the correlation between SFRP-1 and SFRP-2 hypermethylation and immunophenotypic features of leukemic blast cells. The result found significant association between hypermethylation of SFRP-1 gene and elevated expression of CD34 (P = 0.045) and CD14 (P = 0.046). While no significant association was found between SFRP-1 hypermethylation and other leukemic blast cell antigenes. SFRP-2 gene hypermethylation had no association with immunophenotypic features of leukemic blast cells. These findings are compatible with results of a study done by Hou et al. They demonstrated that there was a significant association between SFRP-1 hypermethylation and increased expression of CD34 and CD14 while there was no correlation between SFRP-2 hypermethylation and any blast cell antigenes [27].
Conclusion
In the present study, it was shown that the methylations of SFRP-1 and SFRP-2 genes contingently take place in all subgroup FAB-AML except subgroup M6 in patients with AML at the time of diagnosis. Also in this study, we did not observe an association between methylation of relevant genes and clinical findings in AML patients such as age, sex, WBC and platelets counts, and complete remission after induction therapy. Therefore, methylation of these genes is not the only factor associated with the disease, but also other molecular events are involved. However, more studies with larger sample size are recommended in order to determine the role of hypermethylation of these genes in the pathogenesis of AML and other hematologic malignancies done.
Acknowledgments
Thanks to the Research Council of Tehran University of Medical Sciences, which has provided funding for this research.
| References | ▴Top |
- Lowenberg B, Downing JR, Burnett A. Acute myeloid leukemia. N Engl J Med. 1999;341(14):1051-1062.
doi pubmed - Parkin D, Whelan S, Ferlay J, Teppo L, Thomas D. Cancer Incidence in Five Continents Vol VIII. IARC Scientific publication No 155. Lyon, IARC Scientific publications; 2002.
- Jost E, Schmid J, Wilop S, Schubert C, Suzuki H, Herman JG, Osieka R, et al. Epigenetic inactivation of secreted Frizzled-related proteins in acute myeloid leukaemia. Br J Haematol. 2008;142(5):745-753.
doi pubmed - Klaus A, Birchmeier W. Wnt signalling and its impact on development and cancer. Nat Rev Cancer. 2008;8(5):387-398.
doi pubmed - Jamieson CH, Ailles LE, Dylla SJ, Muijtjens M, Jones C, Zehnder JL, Gotlib J, et al. Granulocyte-macrophage progenitors as candidate leukemic stem cells in blast-crisis CML. N Engl J Med. 2004;351(7):657-667.
doi pubmed - Paul S, Dey A. Wnt signaling and cancer development: therapeutic implication. Neoplasma. 2008;55(3):165-176.
pubmed - Jones SE, Jomary C. Secreted Frizzled-related proteins: searching for relationships and patterns. Bioessays. 2002;24(9):811-820.
doi pubmed - Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781-810.
doi pubmed - Marsit CJ, Karagas MR, Andrew A, Liu M, Danaee H, Schned AR, Nelson HH, et al. Epigenetic inactivation of SFRP genes and TP53 alteration act jointly as markers of invasive bladder cancer. Cancer Res. 2005;65(16):7081-7085.
doi pubmed - Bovolenta P, Esteve P, Ruiz JM, Cisneros E, Lopez-Rios J. Beyond Wnt inhibition: new functions of secreted Frizzled-related proteins in development and disease. J Cell Sci. 2008;121(Pt 6):737-746.
doi pubmed - Mikesch JH, Steffen B, Berdel WE, Serve H, Muller-Tidow C. The emerging role of Wnt signaling in the pathogenesis of acute myeloid leukemia. Leukemia. 2007;21(8):1638-1647.
doi pubmed - Wang H, Fan R, Wang XQ, Wu DP, Lin GW, Xu Y, Li WY. Methylation of Wnt antagonist genes: a useful prognostic marker for myelodysplastic syndrome. Ann Hematol. 2013;92(2):199-209.
doi pubmed - Huang ZH, Li LH, Yang F, Wang JF. Detection of aberrant methylation in fecal DNA as a molecular screening tool for colorectal cancer and precancerous lesions. World J Gastroenterol. 2007;13(6):950-954.
pubmed - Zou H, Molina JR, Harrington JJ, Osborn NK, Klatt KK, Romero Y, Burgart LJ, et al. Aberrant methylation of secreted frizzled-related protein genes in esophageal adenocarcinoma and Barrett's esophagus. Int J Cancer. 2005;116(4):584-591.
doi pubmed - Huang J, Zhang YL, Teng XM, Lin Y, Zheng DL, Yang PY, Han ZG. Down-regulation of SFRP1 as a putative tumor suppressor gene can contribute to human hepatocellular carcinoma. BMC Cancer. 2007;7:126.
doi pubmed - Fodde R, Smits R, Clevers H. APC, signal transduction and genetic instability in colorectal cancer. Nat Rev Cancer. 2001;1(1):55-67.
doi pubmed - Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429(6990):457-463.
doi pubmed - Nakamoto D, Yamamoto N, Takagi R, Katakura A, Mizoe JE, Shibahara T. Detection of microsatellite alterations in plasma DNA of malignant mucosal melanoma using whole genome amplification. Bull Tokyo Dent Coll. 2008;49(2):77-87.
doi pubmed - Veeck J, Bektas N, Hartmann A, Kristiansen G, Heindrichs U, Knuchel R, Dahl E. Wnt signalling in human breast cancer: expression of the putative Wnt inhibitor Dickkopf-3 (DKK3) is frequently suppressed by promoter hypermethylation in mammary tumours. Breast Cancer Res. 2008;10(5):R82.
doi pubmed - Veeck J, Geisler C, Noetzel E, Alkaya S, Hartmann A, Knuchel R, Dahl E. Epigenetic inactivation of the secreted frizzled-related protein-5 (SFRP5) gene in human breast cancer is associated with unfavorable prognosis. Carcinogenesis. 2008;29(5):991-998.
doi pubmed - Cooper SJ, von Roemeling CA, Kang KH, Marlow LA, Grebe SK, Menefee ME, Tun HW, et al. Reexpression of tumor suppressor, sFRP1, leads to antitumor synergy of combined HDAC and methyltransferase inhibitors in chemoresistant cancers. Mol Cancer Ther. 2012;11(10):2105-2115.
doi pubmed - Suzuki H, Gabrielson E, Chen W, Anbazhagan R, van Engeland M, Weijenberg MP, Herman JG, et al. A genomic screen for genes upregulated by demethylation and histone deacetylase inhibition in human colorectal cancer. Nat Genet. 2002;31(2):141-149.
doi pubmed - Fukui T, Kondo M, Ito G, Maeda O, Sato N, Yoshioka H, Yokoi K, et al. Transcriptional silencing of secreted frizzled related protein 1 (SFRP 1) by promoter hypermethylation in non-small-cell lung cancer. Oncogene. 2005;24(41):6323-6327.
doi pubmed - Qi J, Zhu YQ, Luo J, Tao WH. Hypermethylation and expression regulation of secreted frizzled-related protein genes in colorectal tumor. World J Gastroenterol. 2006;12(44):7113-7117.
pubmed - Muller HM, Oberwalder M, Fiegl H, Morandell M, Goebel G, Zitt M, Muhlthaler M, et al. Methylation changes in faecal DNA: a marker for colorectal cancer screening? Lancet. 2004;363(9417):1283-1285.
doi - Pehlivan M, Sercan Z, Sercan HO. sFRP1 promoter methylation is associated with persistent Philadelphia chromosome in chronic myeloid leukemia. Leuk Res. 2009;33(8):1062-1067.
doi pubmed - Hou HA, Kuo YY, Liu CY, Lee MC, Tang JL, Chen CY, Chou WC, et al. Distinct association between aberrant methylation of Wnt inhibitors and genetic alterations in acute myeloid leukaemia. Br J Cancer. 2011;105(12):1927-1933.
doi pubmed - Mizuki M, Schwable J, Steur C, Choudhary C, Agrawal S, Sargin B, Steffen B, et al. Suppression of myeloid transcription factors and induction of STAT response genes by AML-specific Flt3 mutations. Blood. 2003;101(8):3164-3173.
doi pubmed - Tickenbrock L, Schwable J, Wiedehage M, Steffen B, Sargin B, Choudhary C, Brandts C, et al. Flt3 tandem duplication mutations cooperate with Wnt signaling in leukemic signal transduction. Blood. 2005;105(9):3699-3706.
doi pubmed
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Journal of Hematology is published by Elmer Press Inc.


